Using Taxon-Environment Relationships: Standardize Taxonomy
Standardize Taxonomy
Naming conventions for the taxonomic hierarchy of different organisms can vary substantially between different datasets. However, to accurately compute inferences, it is critical that all taxonomic identifications are consistent with one another. This step standardizes the taxonomy of your biological data to the Integrated Taxonomic Information System (ITIS), which was developed by a partnership of US federal agencies.
Instructions for loading your data file and reconciling observations with ITIS are provided below.
- Load inference library into your current R workspace.
Type at the R prompt:
library(bio.infer)
- Format and load local benthic macroinvertebrate observation data.
Three columns in the following order are required: (1) Sample identifier, (2) taxon name, and (3) taxon abundance.
Example:
SITE.ID TAXANAME ABUNDANCE A01 SWELTSA SP. 1 A01 HYDROPSYCHIDAE 5 . . . . . . A02 BAETIS SP. 50 Files should be formatted as tab-delimited text.
Type at the R prompt:
bcnt <- read.delim("filename")
This command reads the tab-delimited file specified by filename and saves the results in a data frame called bcnt.
-
Load ITIS database.
Type at the R prompt:
data(itis.ttable)
-
Run the R script that reconciles local taxon names with ITIS.
Type at the R prompt:
bcnt.tax <- get.taxonomic(bcnt, itis.ttable)
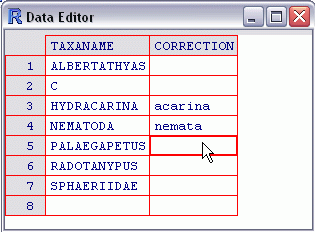 This command runs the R script get.taxonomic and stores the results in the data frame bcnt.tax. Two data sets are required as input to the script, the local benthic count file (bcnt) and the full ITIS taxonomy table (itis.ttable).
This command runs the R script get.taxonomic and stores the results in the data frame bcnt.tax. Two data sets are required as input to the script, the local benthic count file (bcnt) and the full ITIS taxonomy table (itis.ttable).The script attempts to match taxa listed in the local benthic count file with taxon names supplied in ITIS. Usually, a limited number of taxa from the local data cannot be matched. For these taxa, the script provides the user an opportunity to correct any taxa that may be misspelled or that may have a synonym that is recognized by ITIS.
Enter corrected taxon names as shown above, and close the window when finished. Taxon names for which corrections are not known can be left blank.
A summary table of the full taxonomic hierarchy of each taxon in the local benthic file is provide in sum.tax.table.txt. This file can be reviewed using any spreadsheet or text viewer.
