EPA Technology Available for Licensing: Viral-Based Real-Time Quantitative PCR Test for Human Fecal Contamination
Inventors: Kyle Bibby, PhD, Orin C. Shanks, PhD, and Elyse Stachler, MS Inventors (left to right): Kyle Bibby, PhD, Orin C. Shanks, PhD, and Elyse Stachler, MS
Inventors (left to right): Kyle Bibby, PhD, Orin C. Shanks, PhD, and Elyse Stachler, MS
Owned by EPA and the University of Pittsburgh
US Patent Application #15/530,122
Human fecal contamination of clean water sources is a major contributor to the spread of disease worldwide. To monitor and manage this threat, the United States Environmental Protection Agency (EPA), World Health Organization, and European Union rely on bacterial fecal indicator tests, but current methods lack specificity for discriminating human waste from other animal pollution sources, which limits effective handling. Currently recommended fecal indicators also target bacteria, even though viruses are reported to be the dominant cause of waterborne disease. In collaboration with the EPA, researchers at the University of Pittsburgh have developed novel real-time quantitative PCR-based assays that detect a common virus – Cross-Assembly phage (CrAssphage) – in the human gut. This technology is highly sensitive and specific to human waste. When there is a need to differentiate between human and non-human fecal sources of water pollution, CrAssphage could be a critical tool.
Technology Description
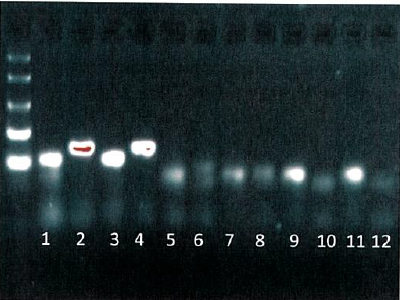 PCR products from example crAssphage primers. Wells 1-4 are raw sewage samples, 5-8 are animal waste samples, and 9-12 are controls.CrAssphage is a recently-discovered bacteriophage that is highly associated with human gut microbiota. The researchers developed a series of PCR primers targeting the CrAssphage genome and then screened them to identify the most sensitive and specific genetic markers for human fecal pollution. Optimal primer sets were adapted to a real-time quantitative PCR platform to allow measurement of human fecal waste concentrations in unknown test samples. Genetic markers were highly abundant in raw sewage samples from across the United States and exhibited high specificity when tested against an animal fecal reference library.
PCR products from example crAssphage primers. Wells 1-4 are raw sewage samples, 5-8 are animal waste samples, and 9-12 are controls.CrAssphage is a recently-discovered bacteriophage that is highly associated with human gut microbiota. The researchers developed a series of PCR primers targeting the CrAssphage genome and then screened them to identify the most sensitive and specific genetic markers for human fecal pollution. Optimal primer sets were adapted to a real-time quantitative PCR platform to allow measurement of human fecal waste concentrations in unknown test samples. Genetic markers were highly abundant in raw sewage samples from across the United States and exhibited high specificity when tested against an animal fecal reference library.
Advantages
- Virus-based technology
- Highly specific to human waste (98.6%)
- Highly sensitive (100%)
Stage of Development
Tested in vitro
IP Status
US non-provisional patent filed December 5, 2016.
Serial number 15/530,122.
Applications
- Waste water treatment and disinfection evaluation
- Waste water discharge permitting
- Watershed management where human fecal pollution is a probable source of contamination
Publications
- Stachler E, Kelty C, Sivaganesan M, Li X, Bibby K, Shanks OC (2017). Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. & Tech., 51:9146-9154.
- Shanks OC, Kelty CA, Oshiro R, Haugland RA, Madi T, Brooks L, Field KG, and Sivaganesan M (2016). Data acceptance criteria for standardized human-assoicated fecal source identification quantitative real-time PCR methods. Appl. Environ. Microbiol, 82: 2773-2782.
- Stachler E, Bibby K (2014). Metagenomic evaluation of highly abundant human gut bacteriophage CrAssphage for source tracking of human fecal pollution. Environmental Science and Technology Letters, 1: 405-409.
- Green H, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walter WQ, Knight R, Kelty CA, and Shanks OC (2014). Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol, 80: 3086-3094.
- Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA (2009). Quantitative PCR for Genetic Markers of Human Fecal Pollution. Appl. Environ. Microbiol, 75(17): 5507-5513.
